The goal of this notebook is to code a decision tree classifier that can be used with the following API:
df = pd.read_csv("data.csv")
train_df, test_df = train_test_split(df, test_size=0.2)
tree = decision_tree_algorithm(train_df)
accuracy = calculate_accuracy(test_df, tree)
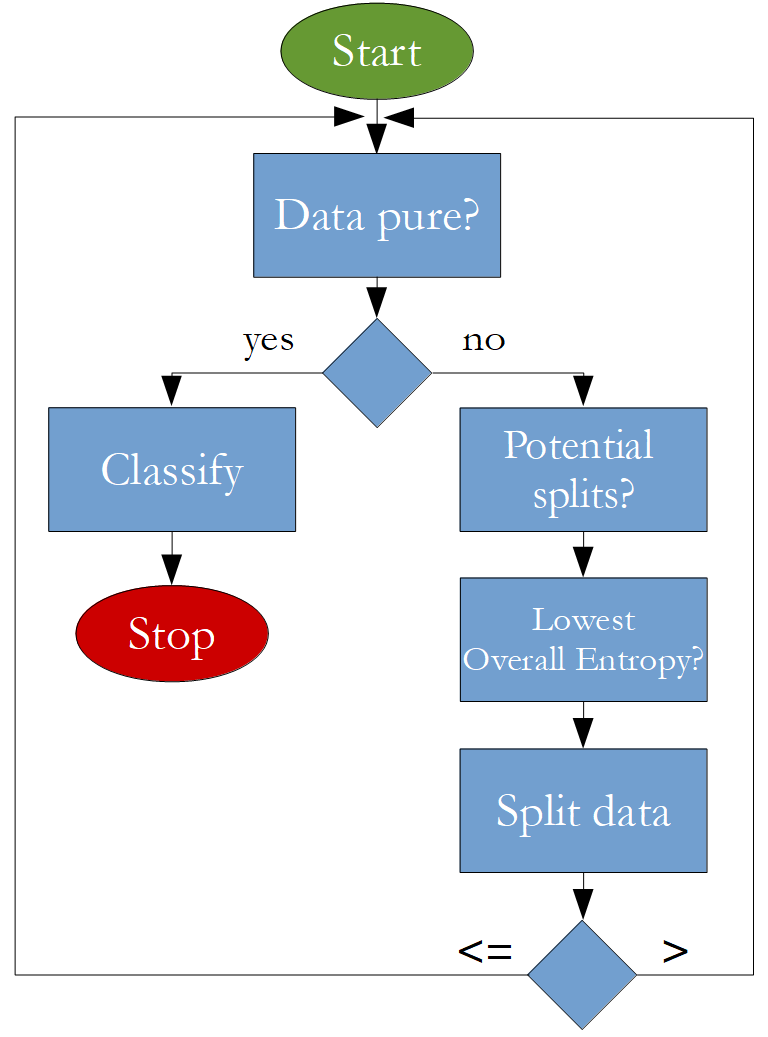
The algorithm that is going to be implemented looks like this:
Import Statements¶
In [1]:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import random
from pprint import pprint
In [2]:
%matplotlib inline
sns.set_style("darkgrid")
Load and Prepare Data¶
Format of the data¶
- the last column of the data frame must contain the label and it must also be called "label"
- there should be no missing values in the data frame
In [3]:
df = pd.read_csv("../data/Iris.csv")
df = df.drop("Id", axis=1)
df = df.rename(columns={"species": "label"})
In [4]:
df.head()
Out[4]:
Train-Test-Split¶
In [5]:
def train_test_split(df, test_size):
if isinstance(test_size, float):
test_size = round(test_size * len(df))
indices = df.index.tolist()
test_indices = random.sample(population=indices, k=test_size)
test_df = df.loc[test_indices]
train_df = df.drop(test_indices)
return train_df, test_df
In [6]:
random.seed(0)
train_df, test_df = train_test_split(df, test_size=20)
Helper Functions¶
The helper functions operate on a NumPy 2d-array. Therefore, let’s create a variable called “data” to see what we will be working with.
In [7]:
data = train_df.values
data[:5]
Out[7]:
Data pure?¶
In [8]:
def check_purity(data):
label_column = data[:, -1]
unique_classes = np.unique(label_column)
if len(unique_classes) == 1:
return True
else:
return False
Classify¶
In [9]:
def classify_data(data):
label_column = data[:, -1]
unique_classes, counts_unique_classes = np.unique(label_column, return_counts=True)
index = counts_unique_classes.argmax()
classification = unique_classes[index]
return classification
Potential splits?¶
In [10]:
def get_potential_splits(data):
potential_splits = {}
_, n_columns = data.shape
for column_index in range(n_columns - 1): # excluding the last column which is the label
potential_splits[column_index] = []
values = data[:, column_index]
unique_values = np.unique(values)
for index in range(len(unique_values)):
if index != 0:
current_value = unique_values[index]
previous_value = unique_values[index - 1]
potential_split = (current_value + previous_value) / 2
potential_splits[column_index].append(potential_split)
return potential_splits
Split Data¶
In [11]:
def split_data(data, split_column, split_value):
split_column_values = data[:, split_column]
data_below = data[split_column_values <= split_value]
data_above = data[split_column_values > split_value]
return data_below, data_above
Lowest Overall Entropy?¶
In [12]:
def calculate_entropy(data):
label_column = data[:, -1]
_, counts = np.unique(label_column, return_counts=True)
probabilities = counts / counts.sum()
entropy = sum(probabilities * -np.log2(probabilities))
return entropy
In [13]:
def calculate_overall_entropy(data_below, data_above):
n = len(data_below) + len(data_above)
p_data_below = len(data_below) / n
p_data_above = len(data_above) / n
overall_entropy = (p_data_below * calculate_entropy(data_below)
+ p_data_above * calculate_entropy(data_above))
return overall_entropy
In [14]:
def determine_best_split(data, potential_splits):
overall_entropy = 9999
for column_index in potential_splits:
for value in potential_splits[column_index]:
data_below, data_above = split_data(data, split_column=column_index, split_value=value)
current_overall_entropy = calculate_overall_entropy(data_below, data_above)
if current_overall_entropy <= overall_entropy:
overall_entropy = current_overall_entropy
best_split_column = column_index
best_split_value = value
return best_split_column, best_split_value
Decision Tree Algorithm¶
Representation of the Decision Tree¶
In [15]:
sub_tree = {"question": ["yes_answer",
"no_answer"]}
In [16]:
example_tree = {"petal_width <= 0.8": ["Iris-setosa",
{"petal_width <= 1.65": [{"petal_length <= 4.9": ["Iris-versicolor",
"Iris-virginica"]},
"Iris-virginica"]}]}
Algorithm¶
In [17]:
def decision_tree_algorithm(df, counter=0):
# data preparations
if counter == 0:
data = df.values
else:
data = df
# base cases
if check_purity(data):
classification = classify_data(data)
return classification
# recursive part
else:
counter += 1
# helper functions
potential_splits = get_potential_splits(data)
split_column, split_value = determine_best_split(data, potential_splits)
data_below, data_above = split_data(data, split_column, split_value)
# instantiate sub-tree
question = "{} <= {}".format(split_column, split_value)
sub_tree = {question: []}
# find answers (recursion)
yes_answer = decision_tree_algorithm(data_below, counter)
no_answer = decision_tree_algorithm(data_above, counter)
sub_tree[question].append(yes_answer)
sub_tree[question].append(no_answer)
return sub_tree
In [18]:
tree = decision_tree_algorithm(train_df)
pprint(tree)
In [ ]: